SARS-CoV-2, the virus that causes COVID-19, is constantly changing and accumulating mutations in its genetic code over time. New variants of SARS-CoV-2 are expected to continue to emerge. Some variants will emerge and disappear, while others will emerge and continue to spread and may replace previous variants.
To identify and track SARS-CoV-2 variants, CDC uses genomic surveillance. CDC’s national genomic surveillance system collects SARS-CoV-2 specimens for sequencing through the National SARS-CoV-2 Strain Surveillance (NS3) program, as well as SARS-CoV-2 sequences generated by commercial or academic laboratories contracted by CDC and state or local public health laboratories. Virus genetic sequences are analyzed and classified as a particular variant. The proportion of variants in a population are calculated nationally, by HHS region, and by jurisdiction. The thousands of sequences analyzed every week through CDC’s national genomic sequencing and bioinformatics efforts fuel the comprehensive and population-based U.S. surveillance system established to identify and monitor the spread of variants.
Rapid virus genomic sequencing data combined with phenotypic data are further used to determine whether COVID-19 tests, treatments, and vaccines authorized or approved for use in the United States will work against emerging variants.
More About Variants
Understanding Variants
Variants and Genomic Surveillance
About These Data
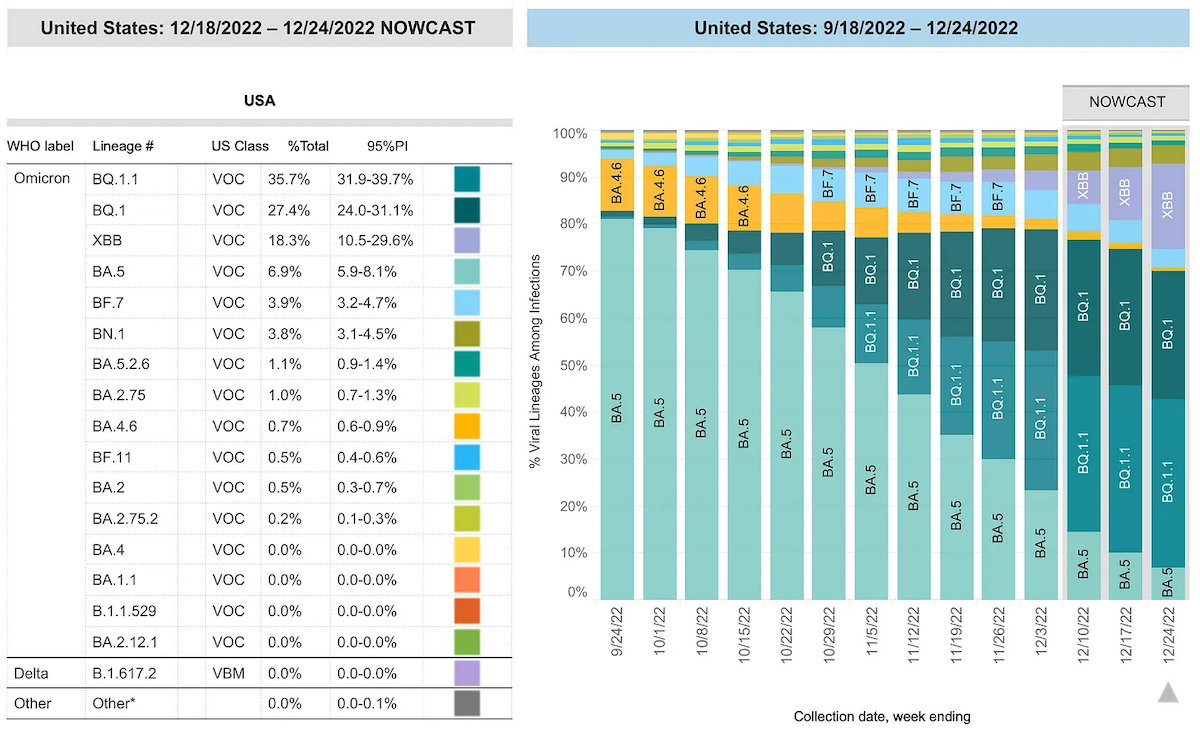
All Variants in the United States
Instructions: Data in the chart and table show the estimated variant proportions for the most common variants and timeframe. The U.S. map shows the estimated weekly proportions of the most common SARS-CoV-2 variants circulating in the United States, divided by HHS regions. Data can be filtered by timeframe (one-week periods), and national or HHS region from the drop-down controls on the top. If a specific timeframe is selected in the chart, the data will change in the table and map to reflect the selected timeframe. If a specific region is selected in the U.S. map, the data will change in the table and map to reflect the selected region. For example, if Region 4 is selected, data will reflect estimates based on reported results from MS, GA, AL, TN, KY, NC, SC, and FL. Data for a specific variant can be highlighted in all figures by selecting it in the “Highlight Variant” box in the left bottom corner of U.S. map, or in the chart, table or piechart in the U.S. map. To see the proportions and their confidence intervals/prediction intervals for all the common variants in the specific timeframe, hover pointer over a bar (timeframe) in the chart. To see the change of the proportion of a variant in different timeframes in a specific region, hover pointer to that variant in the specific region in U.S. map (a table showing proportions of other variants will also show up).
Nowcasting: The default setting for the chart, table, and U.S. map is to display CDC’s Nowcast estimates. Because it can take 2-3 weeks from the time a specimen is collected to when its sequence data is available for analysis, Nowcast is an important tool that can estimate variant proportions for more recent time intervals. Nowcast does not predict future spread of the virus, but it does help estimate current prevalence of variants, based on genomic surveillance data from previous weeks. Estimates of variant proportions for previous weeks may change as more data are reported. Nowcast estimates consistently align with the weighted proportions based on reported sequencing data, which are published 2-3 weeks later.
Weighted Proportions
To provide more representative national, regional, and jurisdiction-level estimates of variant proportions, calculations are included to account for sampling of data over time and across or within states. For example, sequences generated from outbreak investigations are often from a very narrow geographical region (such as a school) and may skew proportions within its larger jurisdiction. Using a survey-design-based approach, CDC uses statistical weights for these estimates that are based on the total number of reverse transcription polymerase chain reaction (RT-PCR) tests and number of SARS-CoV-2-positive RT-PCR test results received, stratified by state, specimen collection date, and by genomic surveillance data source. Variant proportions are estimated based on genomic sequences obtained through CDC (NS3 and CDC-funded sequencing contracts) and tagged baseline surveillance sequencing submitted to public repositories by state, local, academic, or commercial laboratories. Sequences used in this analysis are intended to be a representative sample of all cases during the selected timeframe and location. They may not match cases reported by states, territorial, tribal, and local officials. Estimates of weighted variant proportions are subject to change, as sequence data from specimens previously collected continues to increase over time.
The variant data reported for the jurisdiction-level estimates are limited to those designated as a variant of concern (VOC), variant of interest (VOI), or variant being monitored (VBM) by the U.S. government SARS-CoV-2 Interagency Group (SIG). Differences in the number of SARS-CoV-2 positive RT-PCR tests, sources of sequence data, and number of sequences available during a period affects the degree of certainty in the weighted proportion estimates. Confidence intervals are provided to describe these uncertainties. These data will be updated every Friday.
